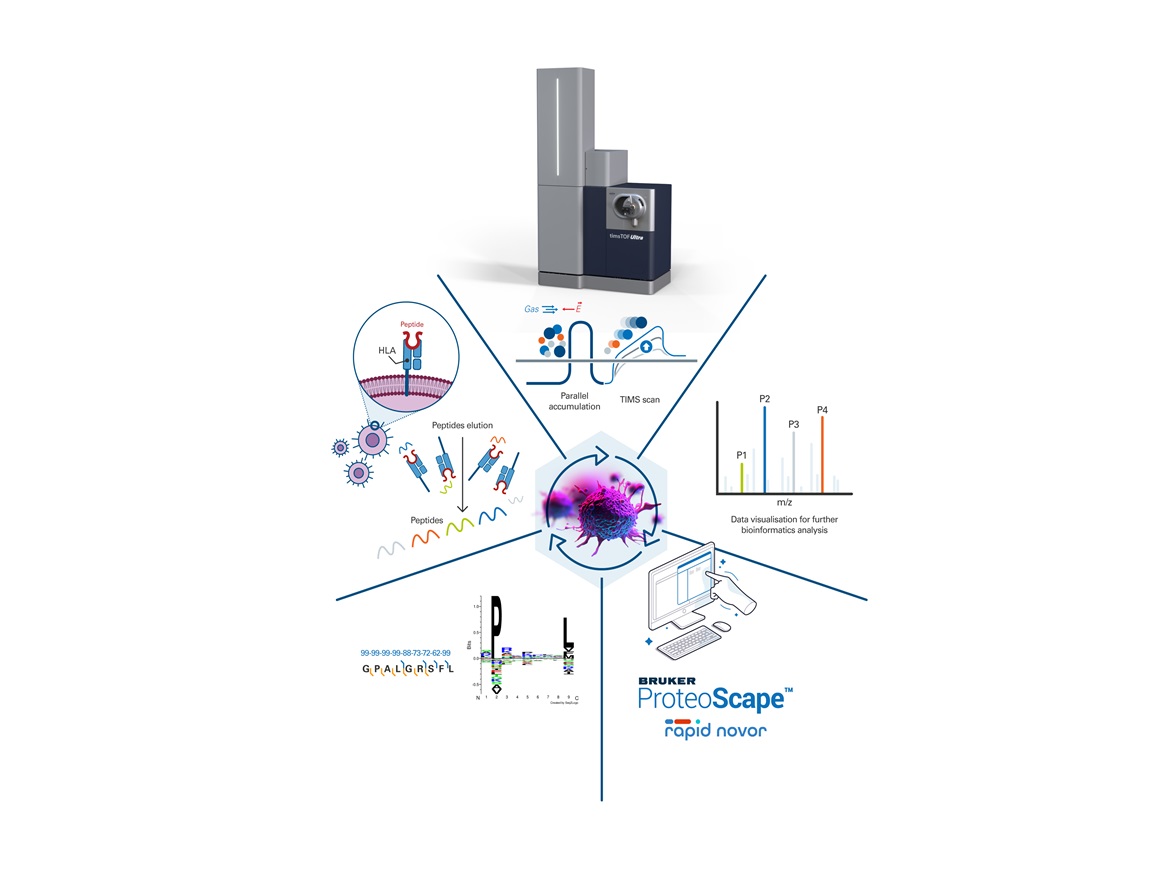
- Novor V2.0 software with novel ML algorithm trained on HLA peptides yields immunopeptidome from de novo peptide sequencing with further improved IDs
- TwinScape ™ digital-twin technology with iRT ™ standards supports highest timsTOF performance with intelligent feedback – well beyond ‘just up-time’
- New glyco-PASEF® method for ultra-high sensitivity 4D glycopeptide analysis
- Spectronaut® 18 software now available for Bruker ProteoScape ™ users
- Biognosys launches high-throughput ENRICHMENT plasma proteomics service for large-scale quantification of ~4.5k proteins with ~55k peptides at 1% FDR specificity
- PreOmics® sample prep tools for proteomics now with >50 BeatBox® placements for fast cell lysis and affordable, high-performance FFPE/FF tissue homogenization
At the 20th US Human Proteome Organization Congress 2024, Bruker Corporation (Nasdaq: BRKR) announced progress in immunopeptidomics, glycoproteomics and other CCS-enabled 4D-proteomics workflows. The rapid advances in deep, at-scale proteomics, glycomics and peptidomics research solutions complement other performance-leading life-science tools for the post-genomic era by Bruker. Together they enable post-genomic molecular and cell biology research, with key insights into disease biology and biomarkers for next-gen molecular diagnostics and drug discovery.
The advances featured at US HUPO include improved machine-learning de novo sequencing (Novor V2.0), a novel high-performance monitoring digital-twin software (TwinScape), the new glyco-PASEFmethod for high-sensitivity glycopeptide analysis, and access to Spectronaut 18 for users of Bruker ProteoScape, a GPU-powered software for real-time 4D proteomics, now with access to library-free searches by Biognosys directDIA ® .
A. Advancing Cancer Immunotherapy Research
Bruker enhances its timsTOF HT and timsTOF Ultra systems with the new software Novor v2.0 for de novo immunopeptidomic profiling. Novor v2.0, developed in collaboration with Rapid Novor Inc., was trained on >1,400,000 spectra mapping to >150,000 HLA peptides for research on small amounts of patient material derived from fine needle biopsies.
Dr. Juliane Walz, Professor of Peptide-based Immunotherapy and Medical Director for Translational Immunology at Universitätsklinikum Tübingen in Germany commented: “Our research focuses on the field of immunology and the development of novel peptide-based immunotherapy concepts for tumor and infectious diseases. Bruker’s trapped ion mobility separation combination with time-of-flight mass spectrometry enables immunopeptidomics with high speed and sensitive detection of HLA-presented peptides. Applying this technology, we have expanded the benign reference immunopeptidome databases with >150,000 HLA peptides. This allows us to confidently define tumor-associated antigens to fast-track immunotherapy development.”
B. Peak timsTOF Performance using TwinScape
TwinScape is a new digital-twin performance monitoring and quality control engine, leveraging iRT peptide standards to support sustained peak performance in sample prep and LC-MS. For synergistic multi-omics studies in the post-genomic era, a combination of depth of coverage, robustness, and scalable quantitation is required. TwinScape enables larger controlled studies and valid cross-lab comparability.
Dr. Anne-Claude Gingras, VP of Research at Sinai Health, Director at Lunenfeld-Tanenbaum Research Institute, and Professor in Molecular Genetics in Toronto, Canada said: “We support a variety of projects in our proteomics facility. It is critical that our instruments are at peak performance and that we always have a pulse on them. The combination of Bruker ProteoScape, TwinScape and the Biognosys iRT kit have become integral for monitoring our instruments to ensure everything is operating up to highest standards. TwinScape is great for an over-time view of QC and system performance. It has been especially useful in pinpointing if there ever is a sample, LC or MS problem.”
C. New glyco-PASEF for Ultra-High Sensitivity 4D-Glycopeptide Analysis
Bruker presents the novel timsTOF glyco-PASEF method with polygon filtering developed with the Heck Group at Utrecht University, the GlycoScape software developed collaboratively with the Van Gool Group at Radboud University, and MSFragger-Glyco software developed by the Nesvizhskii Group at the University of Michigan.
Dr. Albert Heck, Professor of Chemistry and Pharmaceutical Sciences at Utrecht University and Scientific Director of the Netherlands Proteomics Center, noted: “I am pleased with our pioneering work in leveraging the unique strengths of the timsTOF platform for glycoproteomics, now translated to the glyco-PASEF method for research in glycobiology.”
Bruker is now offeringGlycoScape™ real-time glycoproteomic software, developed with Alain van Gool’s team at Radboud University, for early access beta-testing.
Bruker also announces that the MSFragger-Glyco software now supports timsTOF data. Dr. Alexey Nesvizhskii, Professor of Computational Medicine, Bioinformatics and Pathology at University of Michigan, and creator of MSFragger, stated: “My team is gratified by the popularity of MSFragger for ultrafast searches, including the identification of post-translationally modified peptides. Our software suite supports the analysis of PASEF data, and we are delighted that Bruker now offers it with timsTOF instruments. This recognition acknowledges the broad applicability of our tools to timsTOF-generated proteomics data.”
D. Software Advances for dia-PASEF®
The newest release of Bruker ProteoScape makes a Spectronaut 18 module available for dia-PASEF analysis, enabling real-time results using GPU computing, including directDIA for library-free searches. Bruker ProteoScapenow also upgrades TIMS DIA-NN software to version 3.0 with improved quantitation accuracy.